« Previous « Start » Next »
5 Advanced Use of Forward Mode for First Derivatives
We have seen in section
4 how
Mad via
overloaded operations on variables of the
fmad class may calculate
first derivatives. In this section we explore more advanced use of
the
fmad class.
5.1 Accessing The Internal Representation of Derivatives
We saw in section
4 that
fmad returns
multiple derivatives as N-D arrays of one dimension higher than the
corresponding value. Since
Matlab variables are by default
matrices, i.e. arrays of dimension 2, then we see that by
default for multiple directional derivatives
fmad will return 3-D (or higher dimension arrays). Although this
approach is systematic it can be inconvenient. Frequently we require
the Jacobian matrix of a vector valued function. We can trivially
reshape or
squeeze the 3-D array obtained by
fmad
to get the required matrix. However, and as we shall see in
Section
5.3, this approach will not work if we wish
to use
Matlab's sparse matrix class to propagate derivatives. We
should bear in mind that the
derivvec class stores arbitrary
dimension derivative arrays internally as matrices. This
internal representation of the derivatives may be accessed directly
through use of the function
getinternalderivs as the
following example shows.
Example 1
MADEXIntRep1: Internal Representation
This example illustrates MAD's internal representation of derivatives.
See also fmad, MADEXExtRep1, MADEXExtRep2
Contents
-
Define a vector [1;1] with derivatives given by the sparse identity
- Getderivs returns the external form in full storage
- Getinternalderivs returns the internal form
Define a vector [1;1] with derivatives given by the sparse identity
format compact
a=fmad([1;1],speye(2))
value =
1
1
Derivatives
Size = 2 1
No. of derivs = 2
d =
(1,1) 1
d =
(2,1) 1
Getderivs returns the external form in full storage
da=getderivs(a)
da(:,:,1) =
1
0
da(:,:,2) =
0
1
Getinternalderivs returns the internal form
getinternalderivs maintains the sparse storage used internally by MAD's derivvec class.
da=getinternalderivs(a)
da =
(1,1) 1
(2,2) 1
See Section
5.3 for more information on this
topic.
5.2 Preallocation of Matrices/Arrays
It is well-known in
Matlab that a major performance overhead is
incurred if an array is frequently increased in size, or
grown, to accommodate new
data as the following example shows.
Example 2
MADEXPrealloc1: importance of array preallocation
This example illustrates the importance of preallocation of arrays when using MAD.
Contents
-
Simple loop without preallocation
- Simple loop with preallocation
- Fmad loop without preallocation
- Fmad loop with preallocation
Simple loop without preallocation
Because the array a continuously grows, the following loop executes slowly.
format compact
clear a
n=5000;
x=1;
a(1)=x;
tic; for i=1:n; a(i+1)=-a(i);end; t_noprealloc=toc
t_noprealloc =
0.0563
Simple loop with preallocation
To improve performance we preallocate a.
clear a
a=zeros(n,1);
a(1)=x;
tic; for i=1:n;a(i+1)=-a(i);end;t_prealloc=toc
t_prealloc =
0.0089
Fmad loop without preallocation
With an fmad variable performance is even more dramatically reduced when growing an array.
clear a
x=fmad(1,[1 2]);
a(1)=x;
tic;for i=1:n;a(i+1)=-a(i);end;t_fmadnoprealloc=toc
t_fmadnoprealloc =
15.1634
Fmad loop with preallocation
To improve performance we preallocate a.
clear a
a=zeros(n,length(x));
a(1)=x;
tic; for i=1:n;a(i+1)=-a(i);end;t_fmadprealloc=toc
t_fmadprealloc =
4.5518
Note that we have preallocated
a using
zeros(n,length(x)). In
fmad we provide fucntions
length,
size and
numel that return, instead of the expected integers,
fmad object results with zero derivatives. This then allows for them to be used in functions such as
zeros (here) or
ones to preallocate
fmad objects. Also note that it is not possible to change the class of the object that is being
assigned to in
Matlab so if we wish to assign
fmad variables to
part of an matrix/array then that array must already be preallocated to be of
fmad
class. This technique has previously been used by Verma
[
Ver98a, Ver98c] in his ADMAT package.
Example 3
In example 5 the input argument y to the
function will be of fmad class. In the third non-comment line,
dydt = zeros(2*N,size(y,2)); % preallocate dy/dt
of the function
of Figure 1 we see that size(y,2) is used as an argument to
zeros to preallocate storage for the variable dydt.
Since y is of class fmad then so will be the result of the
size function, and hence so will be the result of the
zeros function and hence dydt will be of class
fmad. This will then allow for fmad variables to be assigned to it
and derivatives will be correctly propagated.
We now see how to avoid problem 1 noted in Section
4.7.
Example 4
We make use of length, acting on an fmad variable to preallocate the array y to be of fmad class.
>> x=fmad(1,1);
>> y=zeros(2,length(x(1)));
>> y(1)=x;
5.3 Dynamic Propagation of Sparse Derivatives
Many practical problems either possess sparse Jacobians or are
partially separable so that many intermediate expressions have sparse
local Jacobians. In either case it is possible to take advantage of
this structure when calculating
F'(
x)
V in the cases when
V is sparse. In particular using
V=
In, the identity
matrix, facilitates calculation of the Jacobian
F'(
x). In
order to do this we need to propagate the derivatives as sparse
matrices.
Matlab matrices (though not N-D arrays) may be stored in an intrinsic
sparse format for efficient
handling of sparse
matrices [
GMS91].
Verma's ADMAT tool [
Ver98a] can use
Matlab's sparse matrices to
calculate Jacobians of functions which are defined only by scalars and
vectors. In
fmad careful design [
For06] of the
derivvec class for storage
of derivatives has allowed us to propagate sparse derivatives for
variables of arbitrary dimension (N-D arrays). In this respect
fmad has a similar capability to the Fortran 90 overloading package
ADO1 [
PR98], and the Fortran source-text translation tool
ADIFOR [
BCH+98] which may make use of the SparseLinC
library to form linear combinations of derivative vectors. See [
Gri00, Chap. 6] for a review of this AD topic.
5.3.1 Use of Sparse Derivatives
In order to use sparse derivatives we initialise our independent
variables with a sparse matrix. We use the feature of the
fmad
constructor (section
4.3) that the derivatives supplied must be of the correct
number of elements and in column major order to avoid having to supply
the derivative as an N-D array. As the following example shows,
getinternalderivs must be used to access derivatives in their sparse storage format.
Example 5
Sparse1: Dynamic sparsity for Jacobians with fmad
This example illustrates use of dynamic sparsity when calculating Jacobians with fmad.
Contents
-
A function with sparse Jacobian
- Initialising sparse derivatives
- Using sparse derivatives
- Extracting the derivatives
- Accessing the internal derivative storage
A function with sparse Jacobian
As a simple, small example we take,
y = F(x_1, x_2)=[ x_1^2 + x_2
x_2^2 ]
for which
DF/Dy = [ 2x_1 1
0 2x_2]
and at (x_1,x_2)=(1,2)' we have,
DF/Dy = [ 2 1
0 4]
Initialising sparse derivatives
We set the derivatives using the sparse identity matrix speye. Note that fmad's deriv component stores the derivatives as a derivvec object with sparse storage as indicated by the displaying of only non-zero entries.
x=fmad([1 2],speye(2))
value =
1 2
Derivatives
Size = 1 2
No. of derivs = 2
d =
(1,1) 1
d =
(1,2) 1
Using sparse derivatives
We perform any calculations in the usual way - clearly y has sparse derivatives.
y=[x(1)^2+x(2); x(2)^2]
value =
3
4
Derivatives
Size = 2 1
No. of derivs = 2
d =
(1,1) 2
d =
(1,1) 1
(2,1) 4
Extracting the derivatives
If we use getderivs then, as usual MAD insists on returning a 3_D array and sparsity information is lost.
dy=getderivs(y)
dy=reshape(dy,[2,2])
dy(:,:,1) =
2
0
dy(:,:,2) =
1
4
dy =
2 1
0 4
Accessing the internal derivative storage
By accessing the internal derivative storage used by fmad in its calculation, the sparse Jacobian can be extracted.
J=getinternalderivs(y)
J =
(1,1) 2
(1,2) 1
(2,2) 4
The above example, though trivial, shows how sparse derivatives may be
used in
Mad. Of course there is an overhead associated with the use
of sparse derivatives and the problem must be sufficiently large to
warrant their use.
Example 6
MADEXbrussodeJacSparse: Brusselator Sparse Jacobian
This example shows how to use the fmad class to calculate the Jacobian of the Brusselator problem using sparse propagation of derivatives. The compressed approache of MADEXbrussodeJacComp may be more efficient.
See also: brussode_f, brussode_S, MADEXbrussodeJac, MADEXbrussodeJacComp, brussode
Contents
-
Initialise Variables
- Use fmad with sparse storage to calculate the Jacobian
- Using finite-differencing
- Comparing the AD and FD Jacobian
Initialise Variables
We define the input variables.
N=3;
t=0;
y0=ones(2*N,1);
Use fmad with sparse storage to calculate the Jacobian
We initialise y with the value y0 and derivatives given by the sparse identity matrix of size equal to the length of y0. We may then evaluate the function and extract the value and Jacobian.
y=fmad(y0,speye(length(y0))); % Only significant change from MADEXbrussodeJac
F=brussode_f(t,y,N);
F_spfmad=getvalue(F); % grab value
JF_spfmad=getinternalderivs(F) % grab Jacobian
JF_spfmad =
(1,1) -2.6400
(2,1) 1.0000
(3,1) 0.3200
(1,2) 1.0000
(2,2) -1.6400
(4,2) 0.3200
(1,3) 0.3200
(3,3) -2.6400
(4,3) 1.0000
(5,3) 0.3200
(2,4) 0.3200
(3,4) 1.0000
(4,4) -1.6400
(6,4) 0.3200
(3,5) 0.3200
(5,5) -2.6400
(6,5) 1.0000
(4,6) 0.3200
(5,6) 1.0000
(6,6) -1.6400
Using finite-differencing
y=repmat(y0,[1 2*N+1]);
y(:,2:end)=y(:,2:end)+sqrt(eps)*eye(2*N);
F=brussode_f(t,y,N);
F_fd=F(:,1);
JF_fd=(F(:,2:end)-repmat(F_fd,[1 2*N]))./sqrt(eps)
JF_fd =
-2.6400 1.0000 0.3200 0 0 0
1.0000 -1.6400 0 0.3200 0 0
0.3200 0 -2.6400 1.0000 0.3200 0
0 0.3200 1.0000 -1.6400 0 0.3200
0 0 0.3200 0 -2.6400 1.0000
0 0 0 0.3200 1.0000 -1.6400
Comparing the AD and FD Jacobian
The function values computed are, of course, identical. The Jacobians disagree by about 1e-8, this is due to the truncation error of the finite-difference approximation.
disp(['Function values norm(F_fd-F_spfmad) = ',num2str(norm(F_fd-F_spfmad,inf))])
disp(['Function Jacobian norm(JF_fd-JF_spfmad)= ',num2str(norm(JF_fd-JF_spfmad,inf))])
Function values norm(F_fd-F_spfmad) = 0
Function Jacobian norm(JF_fd-JF_spfmad)= 2.861e-008
5.3.2 User Control of Sparse Derivative Propagation
Some AD tools, for example ADO1 [
PR98],
ADIFOR [
BCH+98], allow for switching from sparse to full
storage data structures for derivatives depending on the number of
non-zeros in the derivative. There may be some user control provided
for this. The sparse matrix implementation of
Matlab [
GMS91] generally does not convert from
sparse to full matrix storage unless a binary operation of a sparse
and full matrix is performed. For example a sparse-full
multiplication results in a full matrix. In the
derivvec arithmetic
used by
fmad we presently provide no facilities for switching from
sparse to full storage once objects have been created. Since MAD version 1.2, the
derivvec class is designed to ensure that once calculations start with sparse derivatives, they continue with sparse derivatives. Any behaviour other than this is a bug and should be reported.
5.4 Sparse Jacobians via Compression
It is well known [
Gri00, Chap. 7] that, subject to a favourable sparsity pattern of the
Jacobian, the number of directional derivatives that need to be
determined in order to form all entries of the Jacobian may be greatly
reduced. This technique, known as
Jacobian (row) compression, is frequently used in finite-difference
approximations of Jacobians [
DS96, Sec. 11.2], [
NW99, p169-173]. Finite-differencing allows
Jacobian-vector products to be approximated and can be used to build
up columns, or linear combinations of columns, of the Jacobian. Use
of sparsity and appropriate coloring algorithms frequently enables
many columns to be approximated from one approximated directional
derivative. This technique is frequently referred to as
sparse finite-differencing however we shall use the term
compressed finite-differencing to avoid confusion with techniques using sparse storage. The forward mode of AD calculates Jacobian-vector
products exactly and, using the same colouring techniques as in compressed
finite-differencing, may therefore calculate many columns of the
Jacobian exactly by propagating just one directional derivative. Thus row compression may also be used by the
fmad class of
Mad.
Coleman and Verma [
CV96] and, independently, Hossain and Steihaug [
HS98] showed
that since reverse mode AD may calculate vector-Jacobian products it
may be used to calculate linear combinations of rows of the Jacobian.
This fact is frequently utilised in optimisation in which an objective
function with single output has its gradient, or row Jacobian
calculated by the propagation of a single adjoint. These authors
showed that by utilising both forward and reverse AD, and specialised
colouring algorithms they could efficiently calculate sparse Jacobians
which could not be calculated by sparse finite-differences. Such techniques are not available in
Mad.
For a
given sparsity pattern the row compression calculation proceeds as follows:
-
Calculate a good coloring from the given sparsity pattern using MADcolor.
- Construct an appropriate seed matrix to initialise derivatives using MADgetseed.
- Calculate the compressed Jacobian by using fmad to propagate derivatives, initialised by the seed matrix, through the function calculation to yield a compressed Jacobian.
- The compressed Jacobian is uncompressed using MADuncompressJac to yield the required sparse Jacobian.
We use the Brusselator example again to illustrate this.
Example 7
MADEXbrussodeJacComp: Brusselator Compressed Jacobian
This example shows how to use the fmad class to calculate the Jacobian of the Brusselator problem using compressed storage. If a good coloring can be found then this approach is usually more efficient than use of sparse or full storage as in MADEXbrussodeJac and MADEXbrussodeJacSparse.
See also: brussode_f, brussode_S, MADEXbrussodeJac, MADEXbrussodeJacSparse, brussode
Contents
-
Initialise variables
- Obtain the sparsity pattern
- Determine the coloring
- Determine the seed matrix
- Calculate the compressed Jacobian using fmad
- Uncompress the Jacobian
- Using finite-differencing
- Comparing the AD and FD Jacobians
- Using compressed finite-differencing
- Comparing the AD and compressed FD Jacobians
Initialise variables
We define the input variables, setting N to 10 for this example.
format compact
N=10;
t=0;
y0=ones(2*N,1);
Obtain the sparsity pattern
The function brussode_S returns the sparsity pattern, the matrix with an entry 1 when the corresponding Jacobian entry is nonzero and entry 0 when the corresponding Jacobian entry is always zero.
sparsity_pattern=brussode_S(N);
figure(1)
spy(sparsity_pattern)
title(['Sparsity pattern for the Brusselator Problem with N=',num2str(N)])
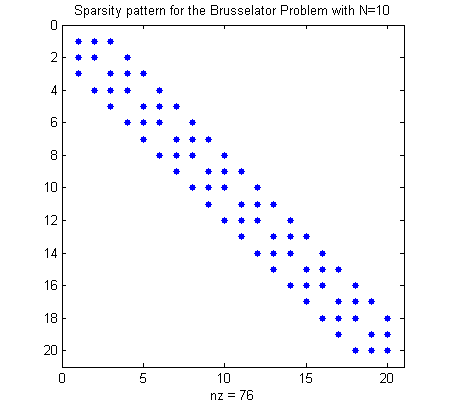
Determine the coloring
The function MADcolor takes the sparsity pattern and determines a color (or group number) for each component of the input variables so that perturbing one component with that color affects different dependent variables to those affected by any other variable in that group.
color_groups=MADcolor(sparsity_pattern);
ncolors=max(color_groups) % number of colors used
ncolors =
4
Determine the seed matrix
The coloring is used by MADgetseed to determine a set of ncolors directions whose directional derivatives may be used to construct all Jacobian entries.
seed=MADgetseed(sparsity_pattern,color_groups);
figure(2);
spy(seed);
title('Sparsity pattern of the seed matrix')
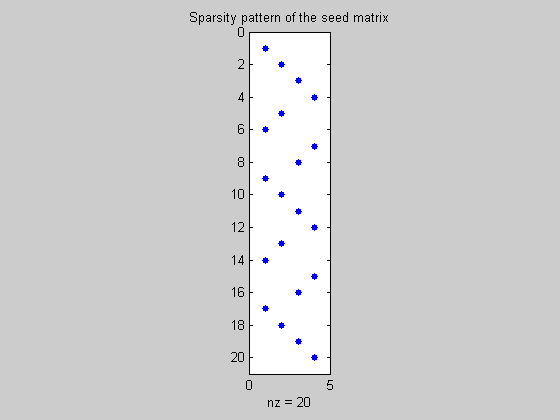
Calculate the compressed Jacobian using fmad
We initialise y with the value y0 and derivatives given by the seed matrix. We then evaluate the function and extract the value and compressed Jacobian.
y=fmad(y0,seed);
F=brussode_f(t,y,N);
F_compfmad=getvalue(F); % grab value
Jcomp=getinternalderivs(F); % grab compressed Jacobian
figure(3);
spy(Jcomp);
title('Sparsity pattern of the compressed Jacobian')
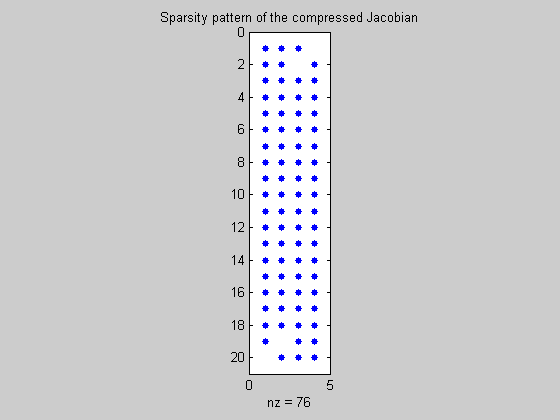
Uncompress the Jacobian
MADuncompressJac is used to extract the Jacobian from its compressed form.
JF_compfmad=MADuncompressJac(Jcomp,sparsity_pattern,color_groups);
figure(4);
spy(JF_compfmad);
title('Sparsity pattern of the calculated Jacobian')
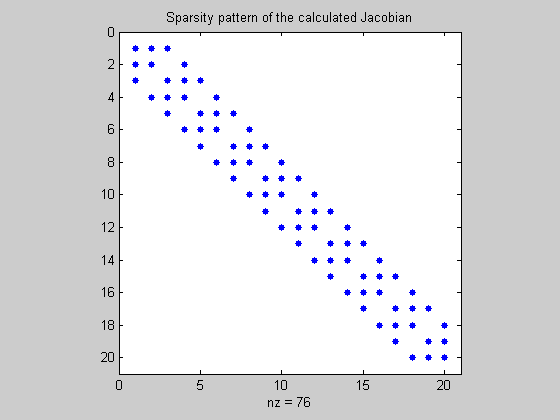
Using finite-differencing
y=repmat(y0,[1 2*N+1]);
y(:,2:end)=y(:,2:end)+sqrt(eps)*eye(2*N);
F=brussode_f(t,y,N);
F_fd=F(:,1);
JF_fd=(F(:,2:end)-repmat(F_fd,[1 2*N]))./sqrt(eps);
Comparing the AD and FD Jacobians
The function values computed are, of course, identical. The Jacobians disagree by about 1e-8, this is due to the truncation error of the finite-difference approximation.
disp(['Function values norm(F_fd-F_compfmad) = ',num2str(norm(F_fd-F_compfmad,inf))])
disp(['Function Jacobian norm(JF_fd-JF_compfmad)= ',num2str(norm(JF_fd-JF_compfmad,inf))])
Function values norm(F_fd-F_compfmad) = 0
Function Jacobian norm(JF_fd-JF_compfmad)= 4.2915e-008
Using compressed finite-differencing
We may also use compression with finite-differencing.
y=repmat(y0,[1 ncolors+1]);
y(:,2:end)=y(:,2:end)+sqrt(eps)*seed;
F=brussode_f(t,y,N);
F_compfd=F(:,1);
Jcomp_fd=(F(:,2:end)-repmat(F_fd,[1 ncolors]))./sqrt(eps);
JF_compfd=MADuncompressJac(Jcomp_fd,sparsity_pattern,color_groups);
Comparing the AD and compressed FD Jacobians
The function values computed are, of course, identical. The Jacobians disagree by about 1e-8, this is due to the truncation error of the finite-difference approximation.
disp(['Function values norm(F_compfd-F_compfmad) = ',num2str(norm(F_compfd-F_compfmad,inf))])
disp(['Function Jacobian norm(JF_compfd-JF_compfmad)= ',num2str(norm(JF_compfd-JF_compfmad,inf))])
Function values norm(F_compfd-F_compfmad) = 0
Function Jacobian norm(JF_compfd-JF_compfmad)= 4.2915e-008
5.5 Differentiating Iteratively Defined Functions
Consider a function
x →
y defined implicitly as a
solution of,
and for which we require ∂
y/∂
x.
Such systems are usually solved via some fixed point iteration of the form,
There have been many publications regarding the most efficient way to
automatically differentiate such a
system [
Azm97, BB98b, Chr94, Chr98] and
reviewed in [
Gri00]. A simple approach termed
piggy-back [
Gri00], is to use forward mode AD through the
iterative process of equation
2. It is necessary to
check for convergence of both the value and derivatives of
y.
Example 8
MADEXBBEx3p1 - Bartholomew-Biggs Example 3.1 [BB98a]
This example demonstrates how to find the derivatives of a function defined by a fixed-point iteration using fmad. The example is taken from (M.C. Bartholomew-Biggs, "Using Forward Accumulation for Automatic Differentiation of Implicitly-Defined Functions", Computational Optimization and Applications 9, 65-84, 1998). The example concerns solution of
with
u1=x1*cos(x2), u2=x1*sin(x2), u3=x3
via the fixed-point iteration,
ynew=yold−g(yold).
See also BBEx3p1_gfunc
Contents
-
Setting up the iteration
- Analytic solution
- Undifferentiated iteration
- Naive differentiation
- Problems with naive differentiation
- Adding a derivative convergence test
- Conclusions
Setting up the iteration
We set the initial value y0, the value of the independent variable x, the tolerance tolg and the maximum number of iterations itmax.
format compact
y0=[0; 0];
x0=[0.05; 1.3; 1]
tolg=1e-8;
itmax=20;
x0 =
0.0500
1.3000
1.0000
Analytic solution
An analytic solution is easily found by setting,
y1=rcosθ, y2=rsinθ,
giving
which may be differentiated to give the required sensitivities.
x=x0;
theta=x(2);
R=sqrt(1+4*x(3)*x(1));
r=(-1+R)/(2*x(3));
y_exact=[r*cos(theta); r*sin(theta)]
dthetadx=[0, 1, 0];
dRdx=[2*x(3)/R, 0, 2*x(1)/R];
drdx=dRdx/(2*x(3))-(-1+R)/(2*x(3)^2)*[0,0,1];
Dy_exact=[-r*sin(theta)*dthetadx+cos(theta)*drdx; ...
r*cos(theta)*dthetadx+sin(theta)*drdx]
y_exact =
0.0128
0.0460
Dy_exact =
0.2442 -0.0460 -0.0006
0.8796 0.0128 -0.0020
Undifferentiated iteration
For the nonlinear, undifferentiated iteration we have:
y=y0;
x=x0;
u=[x(1)*cos(x(2));x(1)*sin(x(2));x(3)];
disp('Itns. |g(y)| ')
for i=1:itmax
g=BBEx3p1_gfunc(y,u);
y=y-g;
disp([num2str(i,'% 4.0f'),' ',num2str(norm(g),'%10.4e')])
if norm(g)<=tolg
break
end
end
y_undif=y
Error_y_undif=norm(y_undif-y_exact,inf)
Itns. |g(y)|
1 5.0000e-002
2 2.5000e-003
3 2.4375e-004
4 2.3216e-005
5 2.2163e-006
6 2.1153e-007
7 2.0189e-008
8 1.9270e-009
y_undif =
0.0128
0.0460
Error_y_undif =
1.6178e-010
Naive differentiation
Simply initialising x with the fmad class gives a naive iteration for which the lack of convergence checks on the derivatives here results in only slight inaccuracy.
y=y0;
x=fmad(x0,eye(length(x0)));
u=[x(1)*cos(x(2));x(1)*sin(x(2));x(3)];
disp('Itns. |g(y)| ')
for i=1:itmax
g=BBEx3p1_gfunc(y,u);
y=y-g;
disp([num2str(i,'% 4.0f'),' ',num2str(norm(g),'%10.4e'),' ',...
num2str(norm(getinternalderivs(g)),'%10.4e')])
if norm(g)<=tolg
break
end
end
y_naive=getvalue(y)
Dy_naive=getinternalderivs(y)
Error_y_naive=norm(y_naive-y_exact,inf)
Error_Dy_naive=norm(Dy_naive-Dy_exact,inf)
Itns. |g(y)|
1 5.0000e-002 1.0000e+000
2 2.5000e-003 1.0003e-001
3 2.4375e-004 1.4508e-002
4 2.3216e-005 1.8246e-003
5 2.2163e-006 2.1665e-004
6 2.1153e-007 2.4729e-005
7 2.0189e-008 2.7469e-006
8 1.9270e-009 2.9909e-007
y_naive =
0.0128
0.0460
Dy_naive =
0.2442 -0.0460 -0.0006
0.8796 0.0128 -0.0020
Error_y_naive =
1.6178e-010
Error_Dy_naive =
2.9189e-008
Problems with naive differentiation
A major problem with naive iteration is if we start close to the solution of the nonlinear problem - then too few differentiated iterations are performed to converge the derivatives. Below only one iteration is performed and we see a large error in the derivatives.
y=y_naive;
x=fmad(x0,eye(length(x0)));
u=[x(1)*cos(x(2));x(1)*sin(x(2));x(3)];
disp('Itns. |g(y)| ')
for i=1:itmax
g=BBEx3p1_gfunc(y,u);
y=y-g;
disp([num2str(i,'% 4.0f'),' ',num2str(norm(g),'%10.4e'),' ',...
num2str(norm(getinternalderivs(g)),'%10.4e')])
if norm(g)<=tolg
break
end
end
y_naive2=getvalue(y)
Dy_naive2=getinternalderivs(y)
Error_y_naive2=norm(y_naive2-y_exact,inf)
Error_Dy_naive2=norm(Dy_naive2-Dy_exact,inf)
Itns. |g(y)|
1 1.8392e-010 1.0000e+000
y_naive2 =
0.0128
0.0460
Dy_naive2 =
0.2675 -0.0482 -0.0006
0.9636 0.0134 -0.0022
Error_y_naive2 =
1.5441e-011
Error_Dy_naive2 =
0.0848
Adding a derivative convergence test
The naive differentiation may be improved by adding a convergence test for the derivatives.
y=y_naive;
x=fmad(x0,eye(length(x0)));
u=[x(1)*cos(x(2));x(1)*sin(x(2));x(3)];
disp('Itns. |g(y)| ')
for i=1:itmax
g=BBEx3p1_gfunc(y,u);
y=y-g;
disp([num2str(i,'% 4.0f'),' ',num2str(norm(g),'%10.4e'),' ',...
num2str(norm(getinternalderivs(g)),'%10.4e')])
if norm(g)<=tolg & norm(getinternalderivs(g))<=tolg % Test added
break
end
end
y_improved=getvalue(y)
Dy_improved=getinternalderivs(y)
Error_y_improved=norm(y_improved-y_exact,inf)
Error_Dy_improved=norm(Dy_improved-Dy_exact,inf)
Itns. |g(y)|
1 1.8392e-010 1.0000e+000
2 1.7554e-011 9.5445e-002
3 1.6755e-012 9.1098e-003
4 1.5992e-013 8.6949e-004
5 1.5264e-014 8.2988e-005
6 1.4594e-015 7.9208e-006
7 1.4091e-016 7.5600e-007
8 1.6042e-017 7.2157e-008
9 4.4703e-019 6.8870e-009
y_improved =
0.0128
0.0460
Dy_improved =
0.2442 -0.0460 -0.0006
0.8796 0.0128 -0.0020
Error_y_improved =
4.1633e-017
Error_Dy_improved =
5.7952e-010
Conclusions
Simple fixed-point iterations may easily be differentiated using MAD's fmad class though users should add convergence tests for derivatives to ensure they, as well as the original nonlinear iteration, have converged. Note that getinternalderivs returns an empty array, whose norm is 0, for variables of class double, so adding such tests does not affect the iteration when used for such variables.
5.6 User Control of Activities/Dependencies
Example 9
MADEXDepend1: User control of activities/dependencies
This example shows how a user may modify their code to render inactive selective variables which MAD automatically assumes are active.
See also: depend, depend2
Contents
-
Function depend
- One independent variable
- Dealing with one dependent variable
Function depend
The function depend defines two variables u,v in terms of independents x,y,
u=x+y, v=−x
Using the fmad class it is trivial to calculate the derivatives du/dx, du/dy, dv/dx, dv/dy. We associate the first directional derivative with the x-derivatives and the second with those of variable y.
format compact
x=fmad(1,[1 0]); y=fmad(2,[0 1]);
[u,v]=depend(x,y);
Du=getinternalderivs(u);
du_dx=Du(1)
du_dy=Du(2)
Dv=getinternalderivs(v);
dv_dx=Dv(1)
dv_dy=Dv(2)
du_dx =
1
du_dy =
1
dv_dx =
-1
dv_dy =
0
One independent variable
Dealing with one independent, say x, is trivial and here requires just one directional derivative.
x=fmad(1,1); y=2;
[u,v]=depend(x,y);
du_dx=getinternalderivs(u)
dv_dx=getinternalderivs(v)
du_dx =
1
dv_dx =
-1
Dealing with one dependent variable
When we are only interested in the derivatives of one dependent variable, say u, then things are more tricky since fmad automatically calculates derivatives of all variables that depend on active inputs. We could, of course, calculate v's derivatives and simply discard them and in many cases (including that here) the performance penalty paid would be insignificant. In some cases a calculation may be significant shortened by ensuring that we calculate derivatives only for those dependent variables we are interested in, or which are involved in calculating those dependents. To illustrate this see function depend2 in which we ensure v is not active by using MAD's getvalue function to ensure that only v's value and not its derivatives, is calculated. u=x+y; v=-getvalue(x); Note that in this case an empty matrix is returned by getinternalderivatives.
x=fmad(1,[1 0]); y=fmad(2,[0 1]);
[u,v]=depend2(x,y);
Du=getinternalderivs(u);
du_dx=Du(1)
du_dy=Du(2)
Dv=getinternalderivs(v)
du_dx =
1
du_dy =
1
Dv =
[]
The last case of just one active output is more difficult to deal with because
derivatives are propagated automatically by
fmad's overloaded library.
Automatically dealing with a restricted number of active outputs
requires a reverse dependency or activity analysis [
Gri00]. This kind of analysis
requires compiler based techniques and is implemented in source
transformation AD tools such as
ADIFOR, TAF/TAMCand Tapenade. In the present
overloading context such an effect can only be achieved as above by the user
carefully ensuring that unnecessary propagation of derivatives
is stopped by judicious use of the
getvalue function to
propagate values and not derivatives.
Of course such coding changes as those shown above, should only be
performed if the programmer is sure they know what they are
doing. Correctly excluding some derivative propagation and hence
calculations may make for considerable run-time savings for large
codes. However incorrect use will result in incorrectly calculated derivative
values.
« Previous « Start » Next »



