A detailed description of the TOMLAB /GENO solver interface is given
below. Also see the M-file help for
.
, are
restricted to be integers.
.
, are
restricted to be integers.
Prob = clsAssign( ... );
| Prob |
Problem description structure. The following fields are used: |
| |
| |
| A |
Linear constraints coefficient matrix. |
| |
| x_L, x_U |
Bounds on variables. |
| |
| b_L, b_U |
Bounds on linear constraints. |
| |
| c_L, c_U |
Bounds on nonlinear constraints. |
| |
| MIP |
Structure with fields defining the integer properties of
the problem. The following fields are used: |
| |
| IntVars |
Vector designating which variables are restricted
to integer values. This field is interpreted
differently depending on the length. |
| |
| |
If length(IntVars) = length(x), it is interpreted as a
zero-one vector where all non-zero elements
indicate integer values only for the corresponding
variable. |
| |
| |
A length less than the problem dimension indicates
that IntVars is a vector of indices for the integer
variables, for example [1 2 3 6 7 12] |
| |
| |
A scalar value K restricts variables x(1) through
x(K) to take integer value only. |
| |
| |
| PriLevOpt |
Print level for the solver. |
| |
| GENO |
Structure with GENO solver specific fields. |
| |
| PrintFile |
Name of file to print progress information and results to. |
| |
| GENO.options |
Structure with special fields for the GENO solver: |
| |
| adj_mode |
This parameter is problem-dependent. It should be set
to 's' for uni-objective optimization problems, or if one seeks a Nash
equilibrium solution of a multi-objective problem; it should be to 'g' in
all other cases. |
| |
Default: None |
| |
| bm_rate |
This parameter is the probability of boundary mutation and
is returned from a simple function. It is problem-dependent but the
default value is usually efficient. |
| |
Default: 0.005 |
| |
| constraints_check |
This parameter allows one to choose whether
or not to display (1) values of the constraints at the end of the program run. |
| |
Default: 0 |
| |
| d_factor |
This is a weighting factor on the direction component
of the differential cross-over operator. The parameter is
problem-dependent but the default range is usually efficient. |
| |
Default: 0.15 - 0.8 |
| |
| m_rate |
This parameter is the probability of ordinary mutation and
is returned from a simple function. It is problem-dependent but the
default value is usually efficient. |
| |
Default: 0.05 |
| |
| maximise |
This parameter is problem-specific: set this to true (1) if the
problem is about maximization. |
| |
Default: 0 |
| |
| p_a_xover |
This parameter is problem-dependent: it specifies the
probability threshold for the arithmetic cross-over operator.
The default value is usually efficient. |
| |
Default: 0.55 |
| |
| p_agents |
This parameter is problem-specific: it declares the number
of sub-objective in a multi-objective problem. |
| |
Default: None |
| |
| p_b_xover |
This parameter is problem-dependent: it specifies the
probability threshold for the boundary cross-over operator. The default
value is usually efficient. |
| |
Default: 0.005 |
| |
| p_d_xover |
This parameter is problem-dependent: it specifies the
probability threshold for the differential cross-over operator. The
default value is usually efficient. |
| |
Default: 0.55 |
| |
| p_eqms |
A constant equal to 1. |
| |
Default: 1 |
| |
| p_h_xover |
This parameter is problem-dependent: it specifies the
probability threshold for the heuristic cross-over operator. The default
value is usually efficient. |
| |
Default: 0.55 |
| |
| p_maxgens |
This parameter specifies the maximum number of generations
that the algorithm will execute. The most efficient value is dependent
on the problem and the population size but it would be safe to
assume that GENO solves most problems within 500 generations. |
| |
Default: None |
| |
| p_mingens |
This parameter should always be 2. |
| |
Default: 2 |
| |
| p_order |
This parameter is problem-specific: it specifies the total
number of variables in the problem. |
| |
Default: None |
| |
| p_plan |
This parameter is problem-specific: it specifies the length
of the control vector along the time dimension. |
| |
Default: None |
| |
| p_popsize |
This parameter specifies the population size. The most
efficient value is problem-dependent but most likely to be within the range shown. |
| |
Default: 10 - 30 |
| |
| p_s_xover |
This parameter is problem-dependent: it specifies the probability
threshold for the simple cross-over operator. The default value is usually efficient. |
| |
Default: 0.55 |
| |
| p_shuffle |
This parameter is problem-dependent: it specifies the probability
threshold for shuffling the population. The default value is usually efficient. |
| |
Default: 0.55 |
| |
| p_u_xover |
This parameter is problem-dependent: it specifies the probability
threshold for the uniform cross-over operator. The default value is usually efficient. |
| |
Default: 0.55 |
| |
| pos_orth |
This parameter is problem-specific: set this to false (0) if the static
constraints are of the 'less than' type. |
| |
Default: 1 |
| |
| quantum_0 |
This parameter is problem-dependent: is specifies the initial size of
quanta. In setting this parameter, the object should be to ensure that the
initial population is sufficiently diverse on all dimensions. In this regard,
a choice of the smaller between 0.1 and 10% of the smallest variable range
is normally efficient. But if one seeks an integer solution, then this
parameter should be set to 1. |
| |
Default: None |
| |
| rand_seed |
This is a seed value for the random number generator. |
| |
Default: None |
| |
| solution_type |
This parameter is problem-dependent: it defines the type of
solution sought. |
| |
Default: None |
| |
| timer |
This parameter declares whether to display (1) GENO's loop time at the
end of the program run. |
| |
Default: 0 |
| |
| vars |
This parameter is problem-specific: it is an 'incidence matrix' that shows
what variables are in each sub-problem of the multi-agent optimization problem. |
| |
Default: None |
| |
| vdu_output |
This parameter declares whether to display (1) progress of the best
chromosome or its fitness. |
| |
Default: 0 |
| |
| view_vars |
In conjunction with vdu_output, this parameter allows one to choose
between viewing (1) the variables in the best chromosome or its fitness. |
| |
Default: 0 |
| |
| Result |
Structure
with result from optimization.
The following fields are set: |
| |
| |
f_k |
Function value at optimum. |
| |
| |
x_k |
Solution vector. |
| |
x_0 |
Initial solution vector. |
| |
| |
c_k |
Nonlinear constraint residuals. |
| |
| |
xState |
State of variables. Free == 0; On lower == 1; On
upper == 2; Fixed == 3; |
| |
bState |
State of linear constraints. Free == 0; Lower ==
1; Upper == 2; Equality == 3; |
| |
cState |
State of nonlinear constraints. Free == 0; Lower
== 1; Upper == 2; Equality == 3; |
| |
| |
Ax |
Values of linear constraints. |
| |
| |
ExitFlag |
Exit status from GENO (TOMLAB standard). 0 = Optimal solution found. |
| |
ExitText |
Exit text from GENO. |
| |
Inform |
GENO information parameter. |
| |
|
0 = Optimal: found an optimal solution. |
| |
|
Other = No optimal solution found. |
| |
| |
FuncEv |
Number of function evaluations. |
| |
ConstrEv |
Number of constraint evaluations. |
| |
QP.B |
Basis vector in TOMLAB QP standard. |
| |
| |
Solver |
Name of the solver (GENO). |
| |
SolverAlgorithm |
Description of the solver. |
| |
| |
GENO |
Subfield with GENO specific results. |
| |
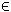 Rn, f(x)
Rn, f(x) 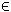 R, A
R, A 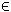 Rm1 × n, bL,bU
Rm1 × n, bL,bU 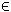 Rm1
and c(x)
Rm1
and c(x) 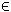 Rm2.
Rm2.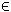 I, the index subset of 1,...,n, are
restricted to be integers.
I, the index subset of 1,...,n, are
restricted to be integers.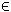 Rn, r(x)
Rn, r(x) 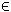 RM, A
RM, A
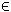 Rm1 × n, bL,bU
Rm1 × n, bL,bU 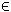 Rm1 and
cL,c(x),cU
Rm1 and
cL,c(x),cU 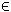 Rm2.
Rm2.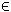 I, the index subset of 1,...,n, are
restricted to be integers.
I, the index subset of 1,...,n, are
restricted to be integers.